Mapping Out Plant Genomes for Human Survival
If you are trying to make sure everyone on earth will have plenty of quality food to eat, you may care to learn how cyanobacteria’s DNA works. The bacteria commonly known as blue-green algae are the origin of chloroplast, and its genomic information provides the fundamental understanding of all plants that depend on photosynthesis to survive. Genomic analysis of such earliest species as well as more recent ones also sheds light on how plants have evolved to overcome ecological challenges over time. Plant genomics is now increasingly gaining the public’s attention as weather patterns grow more unpredictable and extreme, necessitating us to tap into the power of life science to find ways to help plants better adapt to the environment. In 1996, Kazusa DNA Research Institute of Japan successfully sequenced cyanobacteria – which was only the fourth completely sequenced genome in the world and the first in Japan. The Institute has since sequenced the genomes of various crop plants, including strawberries, tomatoes, sweet potatoes and napa cabbage, by collaborating with universities, research institutions and corporations. Dr. Satoshi Tabata, the director of Kazusa DNA Research Institute, who played the central role in the 1996 sequencing of cyanobacteria, will explain how researchers could use genomic data of plants for societal common good.
Ask an Expert: Dr. Satoshi Tabata (Director of Kazusa DNA Research Institute)
Dr. Tabata is an award-winning genome scientist known for his leading role at Kazusa DNA Research Institute in its successful sequencing of cyanobacteria’s complete genome in 1996. He received his bachelor’s of science in biology from Kobe University in 1977 and his doctorate of science from Kyoto University in 1983 before serving as a postdoctoral fellow at the University of California, San Diego in 1982 and as a research assistant at Kyoto University Institute of Chemical Research in 1982. He joined Kazusa DNA Research Institute in 1994 as a researcher, where he has served as director since 2013. He has received the gold medal at Tokyo Techno Forum 1997, a Japanese Society of Plant Physiologists Award and a Kumho Science International Award, both in 2001. For his complete bio, click here.
The “Mutants” on Our Dining Table
Plants cannot move like animals do. That explains why plants have more genes and longer genomes than animals do, according to Dr. Tabata of Kazusa DNA Research Institute.
“Plants have numerous genes that can come in to play in order to help adjust to harsh environments, compensating for their inability to relocate,” Dr. Tabata said. “Animals, on the other hand, have fewer genes, which become reprogrammed in response to environmental changes. So, plants and animals have contrasting survival strategies.”
Parts of the nucleotide sequences in the genome function as genes while the rest are only there to connect the genes without the protein-producing functionality. Plant genomes contain more gene parts and less connection parts than animal genomes, according to Dr. Tabata. Plants’ genomes are also longer.
Many plants also have multiple genomes.
“The human only has one genome comprised of a pair of chromosomes with one side of the pair representing the maternal heredity and the other side paternal. In contrast, some of wheat have sextets of chromosomes, or three genomes. The wheat used to make the bread we eat has three genomes – genomes A, B and D – that are similar to but slightly different from each other,” Dr. Tabata said.
But, why does the genome multiply in plants?
“It probably stems from some sort of mutation that occurs when cells divide. Genome multiplication would kill animals, but plants tend to tolerate it,” Dr. Tabata said.
“Heterosis – crossbred species’ superiority in the growth rate and other features – is also prominent among plants. And we know that genome multiplication causes plants to grow larger. As humans intentionally selected these “mutants” for cultivation, they have become crop plants,” Dr. Tabata said.
These crop plants include sweet potatoes that have a sextet of chromosomes and strawberries that have an octet of chromosomes. Their genomes are longer and more complex than animals’ genomes.
“The genomes of crop plants, in particular, are extremely difficult to analyze for that reason,” Dr. Tabata said.

From the Lab to the Field
Applying the knowledge gained through genomic analysis inside the lab to crop cultivation is a challenging task.
“In research projects, we’ve traditionally used ‘model plants’ that are design-bred for lab uses only. Researchers didn’t know how to take the data from lab experiments conducted in meticulously controlled environments and apply that to the field environment where it would suddenly rain and snow and have diseases striking plants out of the blue,” Dr. Tabata said.
Dr. Tabata pointed out that this gave rise to the question: “Whose job is it to research breeding techniques for crop plants?”
“I thought we must take on the challenge and establish the linkage between genomics and the data from the fields. To do that, we need to be able to scientifically understand field data and develop methodologies for it. We would then record how plants changed under what conditions and evaluate the data. That’s exactly how we are conducting research on crop plants at Kazusa DNA Research Institute,” Dr. Tabata said.
The Japan Science and Technology Agency’s CREST (Create Revolutionary Technological Seeds for Science and Technology Innovation) is currently promoting this initiative of connecting genomics with field data through its program titled “Creation of fundamental technologies contribute to the elucidation and application for the robustness in plants against environmental changes.”
Dr. Tabata believes data and knowledge from plant genomic research should be leveraged to help solve real-world problems, just as human genome research is helping the development of effective treatments for diseases.
“We need to remove some hurdles that are in the way of getting there. I think it’s Kazusa’s job – and the Japanese science community’s job – to figure out how to do just that,” Dr. Tabata said. “And this should be a rather immediate goal and not a distant one. Climate is rapidly changing, and we are seeing different pests and plant diseases migrating from Southeast Asia to as far north as the Kyushu Island. We know we need to quickly develop new varieties that can withstand the increasingly challenging environment.”
Plant Genomics for Common Good
Dr. Tabata said it’s a testament to the advancement of genomic science that people are discussing how to apply the knowledge to cultivation.
“As an undergraduate biology student, I used to wonder how much longer it would take to get the entire genomic picture of a species,” he said. “Back then, biology meant analyzing individual phenomena or individual genes that we knew of and compiling those understandings to create a big picture. Then genomics emerged, and what came along with it was this new approach to biology, in which we first list all substances that are useful to understanding the genomic innerworkings of a species and gather data according to the list to develop the whole picture. That’s what we call data-driven science,” Dr. Tabata said.
For example, researchers would gather and analyze genome-wide data on how different proteins interact with each other, what substances are produced through the metabolic process and what types of chemical chain-reactions those substances have. Kazusa DNA Research Institute is working to expand the databases that allow for such “omics analysis.”
“It's important, Dr. Tabata says, given that genomic databases are generally intended for multidisciplinary uses. What researchers need to remember, however, is that multiple factors play into omics analysis; two identical genetic codes in omics data can mean two different things. This is important, Dr. Tabata says, given that genetic databases generally serve as the platforms for multidisciplinary collaborations. Even if you integrate all the data and find a common gene among different kinds of species, it may function differently in each kind of the species because what caused the gene to be born, such as the environmental factors and other genes, vastly vary in each case,” Dr. Tabata said. “Genomic data provides the foundation for biological research. How to interpret the data and how to layer that knowledge on top of the foundation will be the key in further advancing plant genomics.”
Dr. Tabata has made efforts to advance plant genomics to that end, collaborating for a KAKENHI project called “Platform for Advanced Genome Science (PAGS)” from 2010 to 2014. Using plant genomics for societal common good is important, Dr. Tabata said. After all, the survival of plants means the survival of humans.

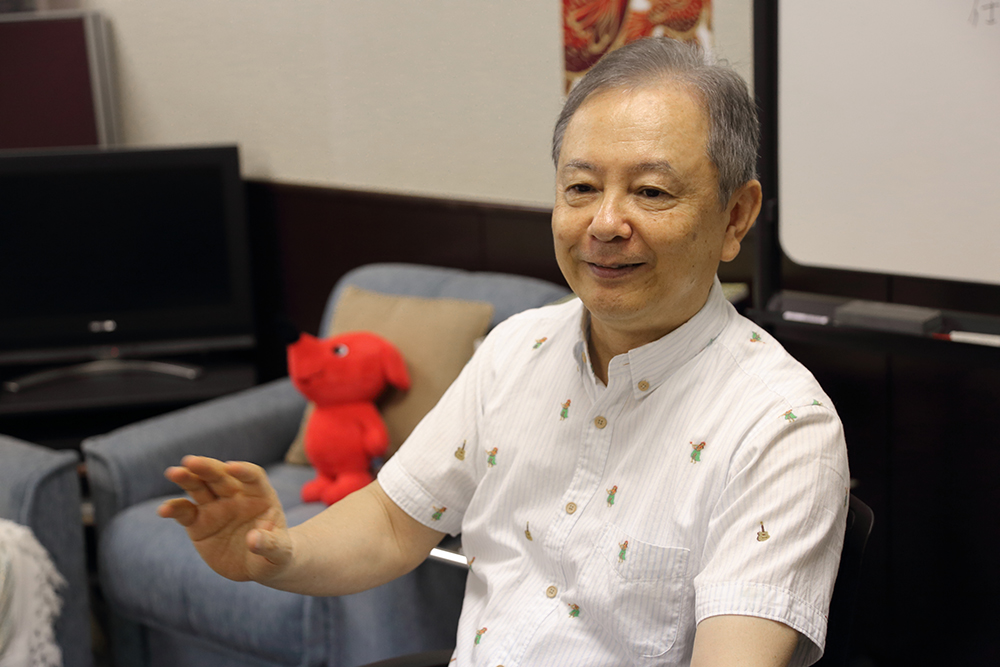
The National BioResource Project (NBRP) contains searchable data on about 250,000 different plant species
Arabidopsis thaliana, rice, wheat, barley, chrysanthemum, morning glory, Lotus japonicus, Glycime max / soja, tomato and algae are among the 30 different bioresources that are registered in the National BioResource Project (NBRP) as strategically important for Japan’s research projects. The NBRP collects, preserves and shares samples of these bioresources and enables searches of data on 250,000 different plant species on its website. Shown here is a screenshot of the rice database.
“Oryzabase” (rice database) – Core operating organization: The National Institute of Genetics)
Interviewer: Rue Ikeya
Photographs: Yuji Iijima unless noted otherwise
Released on: May 31, 2018 (The Japanese version released on Sept. 11, 2017)